We have developed a new PLS method for cell type continuous annotation of single cells, now in preprint!
- Φ-Space addresses numerous challenges faced by state-of-the-art automated annotation methods:
- to identify continuous and out-of-reference cell states,
- to deal with batch effects in reference,
- to utilise bulk references and multi-omic references.
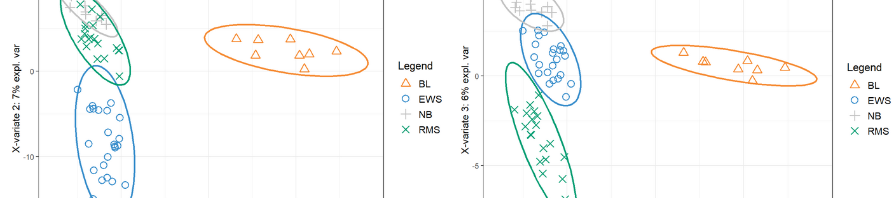
- Φ-Space uses soft classification to phenotype cells on a continuum. The continuous annotation, or phenotype space embedding is then used to reduce the dimensionality of the data for various downstream analyses.
Φ-Space: Continuous phenotyping of single-cell multi-omics data. Jiadong Mao, Yidi Deng, Kim-Anh Lê Cao. bioRxiv 2024.
View this 52min video of Kim-Anh Lê Cao presenting Φ-Space at the WEHI Bioinformatics seminar:
Abstract.
Single-cell multi-omics technologies have empowered increasingly refined characterisa- tion of the heterogeneity of cell populations. Automated cell type annotation methods have been developed to transfer cell type labels from well-annotated reference datasets to emerging query datasets. However, these methods suffer from some common caveats, including the failure to characterise transitional and novel cell states, sensitivity to batch effects and under-utilisation of phenotypic information other than cell types (e.g. sample source and disease conditions).
We developed Φ-Space, a computational framework for the continuous phenotyping of single-cell multi-omics data. In Φ-Space we adopt a highly versatile modelling strategy to continuously characterise query cell identity in a low-dimensional phenotype space, defined by reference phenotypes. The phenotype space embedding enables various downstream analyses, including insightful visualisations, clustering and cell type labelling.
We demonstrate through three case studies that Φ-Space (i) characterises develop- ing and out-of-reference cell states; (ii) is robust against batch effects in both reference and query; (iii) adapts to annotation tasks involving multiple omics types; (iv) over- comes technical differences between reference and query.
The Φ-Space package
Φ-Space is currently not directly available from the mixOmics package, instead it is a separate R package that can be installed from Github.