Note that some webinars listed below describe newly developed statistical methods (e.g. LUPINE, Φ-Space) that are not yet implemented within the mixOmics framework but are available as separate R packages.
- Webinar: Φ-Space ST: a platform-agnostic method to identify cell states in spatial transcriptomics studies
 We have a sequel to Φ-Space, Φ-Space ST developed by Dr Jiadong Mao for spatial transcriptomics studies! We are very excited about these new developments and the potential of Φ-Space for single cell annotation! Φ-Space ST is: Φ-Space ST: a platform-agnostic method to identify cell states in spatial transcriptomics studies. Jiadong Mao, Jarny Choi, Kim-Anh … Continue reading “Webinar: Φ-Space ST: a platform-agnostic method to identify cell states in spatial transcriptomics studies”
We have a sequel to Φ-Space, Φ-Space ST developed by Dr Jiadong Mao for spatial transcriptomics studies! We are very excited about these new developments and the potential of Φ-Space for single cell annotation! Φ-Space ST is: Φ-Space ST: a platform-agnostic method to identify cell states in spatial transcriptomics studies. Jiadong Mao, Jarny Choi, Kim-Anh … Continue reading “Webinar: Φ-Space ST: a platform-agnostic method to identify cell states in spatial transcriptomics studies” - Webinar: PLS methods
 This webinar was presented for a seminar to a group of quantitative researchers (mostly statisticians) at the University of Melbourne. Abstract is below. Topics covered: context of data integration, PCA solved with NIPALS algorithm and SVD, sparse PCA, correlation circle plot interpretation, PLS algorithms and deflation modes, sparse PLS. Technological improvements have allowed for the … Continue reading “Webinar: PLS methods”
This webinar was presented for a seminar to a group of quantitative researchers (mostly statisticians) at the University of Melbourne. Abstract is below. Topics covered: context of data integration, PCA solved with NIPALS algorithm and SVD, sparse PCA, correlation circle plot interpretation, PLS algorithms and deflation modes, sparse PLS. Technological improvements have allowed for the … Continue reading “Webinar: PLS methods” - Webinar: Time-course multi-omics integration
 I presented this talk for a group of statisticians at the Australian National University in Canberra. The abstract is below. Topics covered: linear mixed model splines, multi-omics integration (PLS multiblock), correlation circle plot interpretation, timeOmics. Longitudinal experiments are becoming increasingly popular in omics studies to monitor molecular changes following treatment or during disease progression. Integrating … Continue reading “Webinar: Time-course multi-omics integration”
I presented this talk for a group of statisticians at the Australian National University in Canberra. The abstract is below. Topics covered: linear mixed model splines, multi-omics integration (PLS multiblock), correlation circle plot interpretation, timeOmics. Longitudinal experiments are becoming increasingly popular in omics studies to monitor molecular changes following treatment or during disease progression. Integrating … Continue reading “Webinar: Time-course multi-omics integration” - Webinar: Φ-Space for continuous phenotyping of single-cell multi-omics data
 We have developed a new PLS method for cell type continuous annotation of single cells, now published in Genome Biology! Mao, J., Deng, Y. & Lê Cao, KA. Phi-Space: continuous phenotyping of single-cell multi-omics data. Genome Biol26, 323 (2025). https://doi.org/10.1186/s13059-025-03755-8 View this 50-min video of Kim-Anh Lê Cao presenting Φ-Space at the WEHI Bioinformatics seminar: Abstract. Single-cell multi-omics technologies have … Continue reading “Webinar: Φ-Space for continuous phenotyping of single-cell multi-omics data”
We have developed a new PLS method for cell type continuous annotation of single cells, now published in Genome Biology! Mao, J., Deng, Y. & Lê Cao, KA. Phi-Space: continuous phenotyping of single-cell multi-omics data. Genome Biol26, 323 (2025). https://doi.org/10.1186/s13059-025-03755-8 View this 50-min video of Kim-Anh Lê Cao presenting Φ-Space at the WEHI Bioinformatics seminar: Abstract. Single-cell multi-omics technologies have … Continue reading “Webinar: Φ-Space for continuous phenotyping of single-cell multi-omics data” - Webinar: PCA and PLS-DA
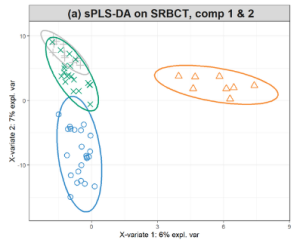 These two recordings were part of a presentation to WEHI for their postgraduate lecture series for a diverse audience. In the PCA presentation (18 min), we explain the concept of linear combination of variables (components) and useful graphical outputs such as correlation circle plots and biplots. In the PLS-DA presentation (7 min), we talk about … Continue reading “Webinar: PCA and PLS-DA”
These two recordings were part of a presentation to WEHI for their postgraduate lecture series for a diverse audience. In the PCA presentation (18 min), we explain the concept of linear combination of variables (components) and useful graphical outputs such as correlation circle plots and biplots. In the PLS-DA presentation (7 min), we talk about … Continue reading “Webinar: PCA and PLS-DA” - Webinar: Microbial network inference for longitudinal microbiome studies with LUPINE
 Our latest method based on PLS to infer microbial networks across time is now in preprint! Microbial network inference for longitudinal microbiome studies with LUPINE. Saritha Kodikara, Kim-Anh Lê Cao. bioRxiv 2024.05.08.593086; accepted in Microbiome. View this 50min video of Dr Saritha Kodikara presenting her method LUPINE: We also have a second video presented by Prof Kim-Anh Lê Cao who sets LUPINE in the … Continue reading “Webinar: Microbial network inference for longitudinal microbiome studies with LUPINE”
Our latest method based on PLS to infer microbial networks across time is now in preprint! Microbial network inference for longitudinal microbiome studies with LUPINE. Saritha Kodikara, Kim-Anh Lê Cao. bioRxiv 2024.05.08.593086; accepted in Microbiome. View this 50min video of Dr Saritha Kodikara presenting her method LUPINE: We also have a second video presented by Prof Kim-Anh Lê Cao who sets LUPINE in the … Continue reading “Webinar: Microbial network inference for longitudinal microbiome studies with LUPINE” - Webinar: mixOmics in 50 minutes
 This latest seminar was hosted by Australian BioCommons / EMBL-ABR / ARDC in March 2024. The latest version includes some recent updates (also covered in the other webinars in more details – check them out!) The slides are opened to the community, but don’t forget to acknowledge the presenter if you are re-using the slides. … Continue reading “Webinar: mixOmics in 50 minutes”
This latest seminar was hosted by Australian BioCommons / EMBL-ABR / ARDC in March 2024. The latest version includes some recent updates (also covered in the other webinars in more details – check them out!) The slides are opened to the community, but don’t forget to acknowledge the presenter if you are re-using the slides. … Continue reading “Webinar: mixOmics in 50 minutes”
