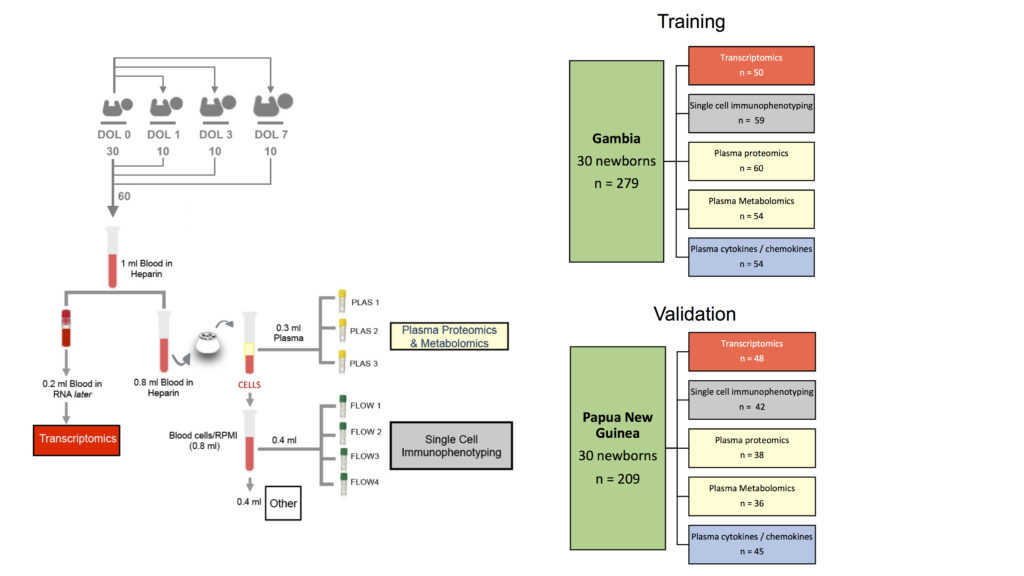
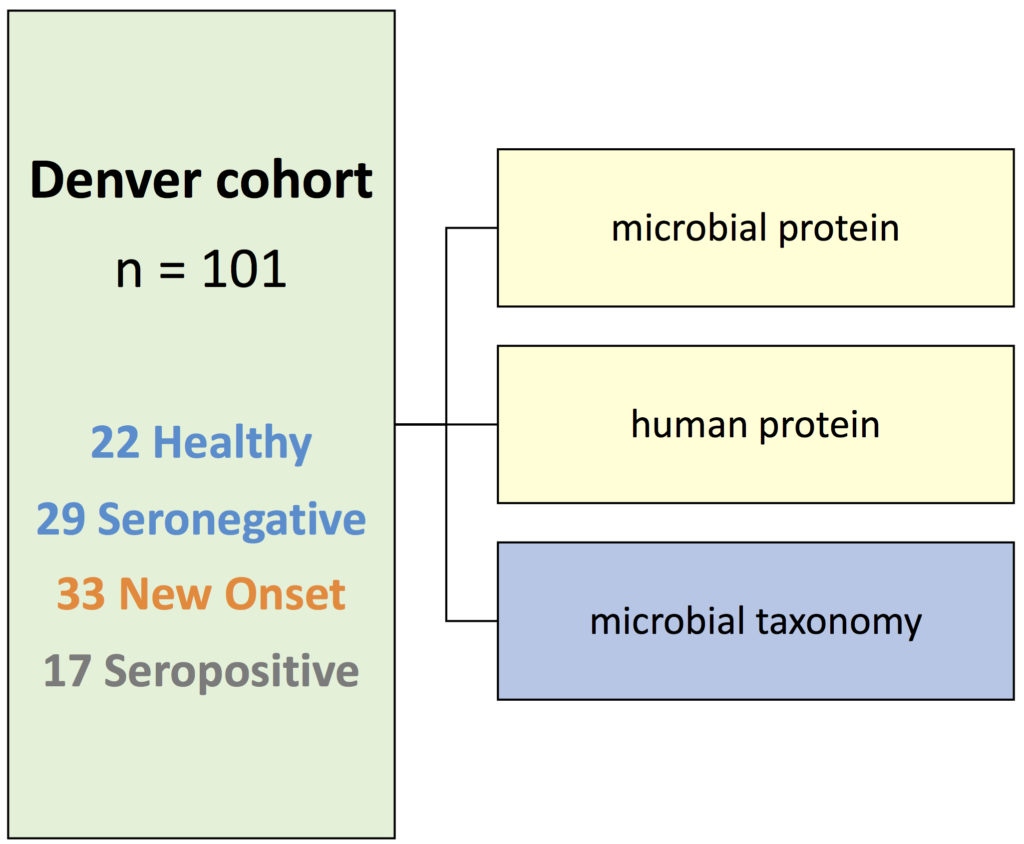
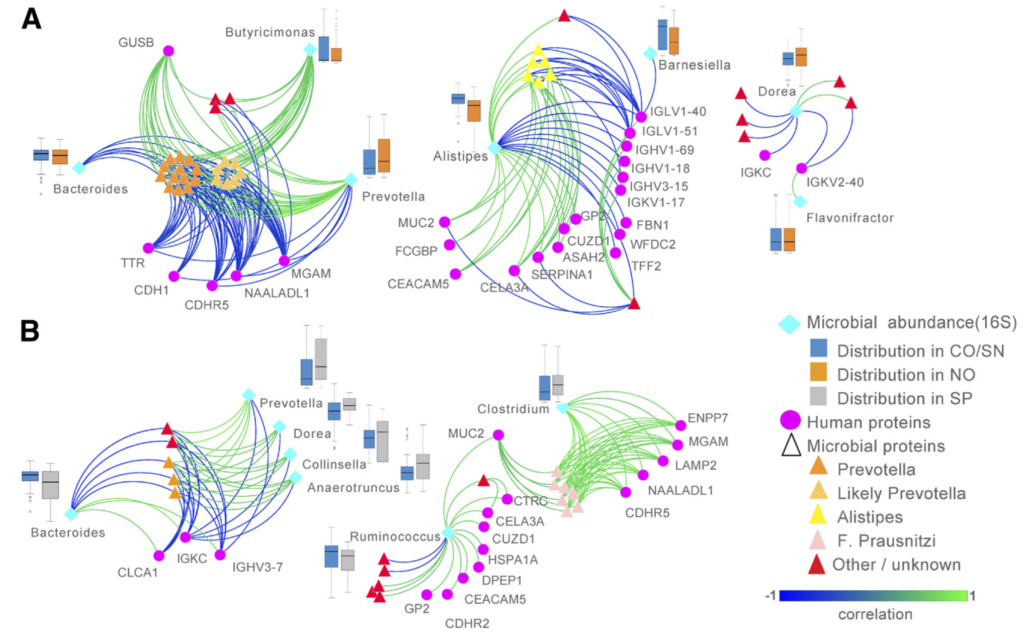
Some feedback from the participants:
‘I liked that simple concepts were explained, to facilitate understanding of the complex ones. It is always nice to recap on the basics’
‘[It] Provided me with novel insights for my research and how to approach and interpret the data from an integrated statistical/computational and biological point of view.’
‘I liked you took some time to explain the concept behind the statistical tool, useful examples to clarify the concepts. After this workshop, it is going to be much easier to make better decisions about future experimental designs (or recommendations for other experiments) and the application of specific statistical tool as MixOmics. I will definitely apply mixMC to the data I am preparing for a publication. I appreciate you granted me the bursary. This workshop has uplifted my microbiome-applied statistical skills.’
‘This was a great workshop and it exceed my expectations’
I could complete all the exercises based on the material provided (which was great, with very clear instructions!). Most importantly, I acquired a better understanding of how I should approach my data in the future when my new project is in a more advanced stage. In other words, I now have a better idea of the packages and tools available and what to focus on in terms of skills development. I am also feeling a lot more confident to interpret complex plots generated in these multivariate analyses and make further decisions based on them and my research question.
Context. Advances in high-throughput technologies have transformed the way we examine molecular information, including microbial communities. However, analytical tool development is critically trailing behind data generation, which hinders the analysis, understanding or integration of microbiome data with other types of molecular data. Data integration adopt a holistic, data-driven and hypothesis-free approach. This new approach is necessary to understand the role of biological systems and posit new hypotheses.
The workshop will introduce concepts of multivariate dimension methods developed in mixOmics for statistical analysis. Our methods make no distributional assumptions, are highly flexible for unsupervised (exploratory), supervised (classification) and integration analyses. Various analytical frameworks will be presented ranging from data exploration, selection of markers, integration with other omics datasets and introduction to time-course analysis.
Each methodology will be illustrated on real biological studies. The third day is ‘BYO data’ day where you can reinforce your learnings on your own study! The workshop is not limited to microbiome data only, as we will cover general omics data integration concepts with appropriate case studies if the need arise.
Instructor: Dr Kim-Anh Lê Cao;Tutor: TBA
Organized and hosted by: West Australian Heath Translational Network (WAHTN) and WA Human Microbiome Collaborating Centre (WAHMCC), Curtin University.

Fees for 3 days are AUD450+GST for RHD students, AUD750+GST for research non-profit organisations (Universities and CSIRO) and AUD1200+GST for industry. The West Australian Heath Translational Network generously sponsors registration bursaries ($225 to support 50% of the registration costs) to 4 RHD students. Apply at the EOI survey link below.
Registrations fees include coffee breaks, lunch, lecture notes and electronic material (slides, R code, data).
Registration Express your interest at this survey link. As we have a limited number of participants (30), priority will be given to postgraduate students and early career researchers. EOI for bursaries closes on November 4 2019 5pm AEST, but there are still spots for non bursaries. Results announced to the participants with details for registration.
Location: Forrest Hall, 35 Stirling Highway, Crawley WA 6009, Australia. Google map.
Accommodation: short stay can be booked at Forrest Hall ($120/night)
Contact: mixomics[ at] math.univ-toulouse.fr (for pre-requisite or content)
Prerequisite and requirements We require from the trainees a good working knowledge in R programming (e.g. handling data frame, perform simple calculations and display simple graphical outputs) to fully benefit from the workshop. Participants are requested to bring their own laptop, having installed the software RStudio http://www.rstudio.com/and the R package mixOmics (instructions will be provided prior to the training).
Outline
Day 1 & 2: methods and hands-on. The following broad topics will be covered.
A. Key methodologies in mixOmics and their variants:
- Basic processing of count data (scaling, how to handle compositional data)
- Exploration of one data set and how to estimate missing values
- Identification of a microbial signature to discriminate different treatment groups
- Integration of two data sets and identification of microbial markers
- Introduction to repeated measurements or longitudinal studies analysis
- How to deal with batch effects
- Integration of more than two data sets to identify multi omics signatures (if sufficient interest)
- Integration of independent but related studies (optional)
B. Review on the graphical outputs implemented in mixOmics
- Sample plot representation
- Variable plot representation for data integration
- Other useful graphical outputs
C. Case studies and applications
Several microbiome and omics studies will be analysed using the methods presented above.
Day 3: bring your own data. Participants will be given the opportunity to analyse their own data under the guidance and the advice of the three instructors. Participants can also work in a team. Some data sets will also be provided for those unable to bring their own data.
The following statistical concepts will be introduced: covariance and correlation, multiple linear regression, classification and prediction, cross-validation, selection of markers, penalised regressions. Each methodology will be illustrated on a case study (theory and application will alternate).
Target group The course is intended for microbiologists working in the fields of bioinformatics, computational biology and applied statistics with some statistical knowledge and a good working knowledge in R. It will be particularly useful to those interested in:
- Exploring data sets.
- Selecting molecular / microbial features with methods implementing LASSO-based penalisations.
- Using graphical techniques to better visualise data.
- Understanding and/or applying multivariate projection methodologies to large data sets.
Anticipated learning outcomes After completion of this workshop, participants will be able to
- Understand fundamental principles of multivariate projection-based dimension reduction technique.
- Perform statistical integration and feature selection using recently developed multivariate methodologies.
- Apply those methods to high throughput microbiome studies, including their own studies.