At the end of October 2024 Bioconductor updated to version 3.20, and with it updated to the latest version of mixOmics 6.30.0. You can install the latest version of mixOmics on Bioconductor here. This latest release version of the package runs on R version 4.4 and includes some minor bug fixes and updated code and unit tests. See our Github page for more details on these updates.
Forum
Webinar: PLS methods
This webinar was presented for a seminar to a group of quantitative researchers (mostly statisticians) at the University of Melbourne. Abstract is below.
Topics covered: context of data integration, PCA solved with NIPALS algorithm and SVD, sparse PCA, correlation circle plot interpretation, PLS algorithms and deflation modes, sparse PLS.
Technological improvements have allowed for the collection of data from different types of molecules (e.g. genes, proteins, metabolites, microorganisms) resulting in multiple ‘omics data (e.g. transcriptomics, proteomics, metabolomics, microbiome) measured from the same set N of biospecimens or individuals. In this talk I will introduce the statistical integration of these multi-omics data to shed more light into a biological system.
Integrating data include numerous challenges – data are complex and large, each with few samples (N < 50) and many molecules (P > 10,000), and generated using different technologies. I will present PLS (Partial Least Squares / Projection to Latent Structures developed by Wold in the 1980s) as an algorithm of choice for data integration of small N large P problems. These variants form the basis of our comprehensive mixOmics R package for feature selection, dimension reduction and integration of omics data sets. This talk is targeted at a general audience with background knowledge in statistics and interest in large data
The webinar was re-recorded for the PLS section.
Webinar: Time-course multi-omics integration
I presented this talk for a group of statisticians at the Australian National University in Canberra. The abstract is below.
Topics covered: linear mixed model splines, multi-omics integration (PLS multiblock), correlation circle plot interpretation, timeOmics.
Longitudinal experiments are becoming increasingly popular in omics studies to monitor molecular changes following treatment or during disease progression. Integrating these data sets can give us some mechanistic insights into the different types of omics layers.
However, longitudinal omics data present numerous challenges including a small number of time points that may be unevenly spaced and unmatched between different data types, a small number of individuals, and a high individual variability. While current approaches have focused on differential expression across time or time profile clustering, the modelling of omics time profiles in a multivariate manner is critically lacking to understand longitudinal biological interactions.
I will present a statistical framework, timeOmics, to identify correlated profiles over time and between omics (transcriptomics, metabolomics, microbiome) to give insights into the molecular dynamics of biological systems and discuss future avenues of research in this expanding area.
Some key references
- Straube J, Gorse AD, PROOF Centre of Excellence Team, Huang BE& and Lê Cao K-A& (2015). A linear mixed model spline framework for analysing time course ‘omics’ data. PLoS ONE 10(8): e0134540
- A Bodein, O Chapleur, A Droit, K-A Lê Cao (2019). A Generic Multivariate Framework for the Integration of Microbiome Longitudinal Studies With Other Data Types, Frontiers in Genetics, 10,
- A Bodein, M-P Scott-Boyer, O Perin, K-A Lê Cao, A Droit (2022). timeOmics: an R package for longitudinal multi-omics data integration, Bioinformatics, 38(2)
The timeOmics package
timeOmics is currently not directly available from the mixOmics package, instead it is a separate R package hosted on Bioconductor. See the Bioconductor page for installation instructions.
[closed] Self-paced online course Feb 24 – April 11, 2025
Single and multi-omics analysis and integration with mixOmics
Our registrations are now closed! Fill in this Expression Of Interest for if you missed out, so that we can notify you of new workshops.
This course is designed for:
- Beginners looking for an introduction to mixOmics methods for single- and multi-omics analyses.
- Current mixOmics users who want to deepen their understanding of the mixOmics methods.
- Users who would like more guidance on analyzing their own data (we also provide exemplar datasets).
The workshop is self-paced and spans across 7 weeks. There are 4 Q&A live sessions, and many opportunities to interact with the cohort and your instructor Prof Kim-Anh Lê Cao via Slack. BYO data is encouraged: we provide advice so that you can analyse your own data with mixOmics tools as part of your learning process. A good working knowledge in R programming (e.g. handling data frame, perform simple calculations and display simple graphical outputs) is essential to fully benefit from the course*.
According to our past participants, a time commitment of 5-8h/week was sufficient to feel that they were progressing. Here is some feedback from a previous course.
We provide a certificate of attendance or completion.
Register here, places are limited!
Fees
Research Higher Degree students enrolled at a University: $495 AUD (incl. GST) [discount code: MIXO_RHD]
Staff and members from Universities & Not-for-profit organisations: $825 (incl. GST) [discount code: MIXO_NFP_STAFF]
Other industries: $1320 AUD (incl. GST)
discounts of 5% for a group of 3-9 learners and 10% for 10+ learners, however, this will require a single invoice per group.
These funds go towards the support of a software developer to maintain the package. If you need an invoice, contact Student Support at continuing-education[at]unimelb.edu.au
Teaching Period Dates
- Teaching commences: Monday, 24 Feb 2025, 9:00 am AEST
- Q&A live webinars are scheduled on Thursdays 6pm AEST / 8am CET during the first 4 weeks (27th Feb, 6th, 13th and 20th March).
- An additional session might be added on Fridays 9am AEST ( = Thursdays 2pm PST / 5pm EST / 9pm CET)
- Teaching concludes: Sunday, 23 March 2025, 11:59 pm AEST (after 4 weeks)
- (non marked) Assessment due: Friday 4 April 2025 (2 weeks prep)
- Peer-review of assessment due: Friday 11 April 2025 (1 week prep)
The course is divided into theory (50%) and hands-on practice, with the opportunity to analyse your own data. The exercises and assignments are in R. Participants are encouraged to use RStudio and Rmarkdown (template and R code provided).
*Need an R refresher?
Learners who are not proficient in R do not get the full benenefit of the course (based on their own, honest, feedback!) For those looking for an R refresher well ahead of the course:
- The R cheatsheets for reference: https://iqss.github.io/dss-workshops/R/Rintro/base-r-cheat-sheet.pdf
Webinar: Φ-Space for continuous phenotyping of single-cell multi-omics data
We have developed a new PLS method for cell type continuous annotation of single cells, now published in Genome Biology!
- Φ-Space addresses numerous challenges faced by state-of-the-art automated annotation methods:
- to identify continuous and out-of-reference cell states,
- to deal with batch effects in reference,
- to utilise bulk references and multi-omic references.
- Φ-Space uses soft classification to phenotype cells on a continuum. The continuous annotation, or phenotype space embedding is then used to reduce the dimensionality of the data for various downstream analyses.
Mao, J., Deng, Y. & Lê Cao, KA. Phi-Space: continuous phenotyping of single-cell multi-omics data. Genome Biol26, 323 (2025). https://doi.org/10.1186/s13059-025-03755-8
View this 50-min video of Kim-Anh Lê Cao presenting Φ-Space at the WEHI Bioinformatics seminar:
Abstract.
Single-cell multi-omics technologies have empowered increasingly refined characterisa- tion of the heterogeneity of cell populations. Automated cell type annotation methods have been developed to transfer cell type labels from well-annotated reference datasets to emerging query datasets. However, these methods suffer from some common caveats, including the failure to characterise transitional and novel cell states, sensitivity to batch effects and under-utilisation of phenotypic information other than cell types (e.g. sample source and disease conditions).
We developed Φ-Space, a computational framework for the continuous phenotyping of single-cell multi-omics data. In Φ-Space we adopt a highly versatile modelling strategy to continuously characterise query cell identity in a low-dimensional phenotype space, defined by reference phenotypes. The phenotype space embedding enables various downstream analyses, including insightful visualisations, clustering and cell type labelling.
We demonstrate through three case studies that Φ-Space (i) characterises develop- ing and out-of-reference cell states; (ii) is robust against batch effects in both reference and query; (iii) adapts to annotation tasks involving multiple omics types; (iv) over- comes technical differences between reference and query.
The Φ-Space package
Φ-Space is currently not directly available from the mixOmics package, instead it is a separate R package that can be installed from Github.
Webinar: PCA and PLS-DA
These two recordings were part of a presentation to WEHI for their postgraduate lecture series for a diverse audience.
In the PCA presentation (18 min), we explain the concept of linear combination of variables (components) and useful graphical outputs such as correlation circle plots and biplots.
In the PLS-DA presentation (7 min), we talk about the concept of multivariate signature.
If you want to know more about the actual algorithm under the hood, you can watch this webinar on PLS.
[closed] Self-paced online course Oct 21 – Dec 6 2024
Unfortunately we had to cancel the workshop as we did not receive a sufficient number of participants to justify running the workshop at this time. These workshops involve peer review and a cohort feel to provide the best experience to our learners.
Register your EOI here and we will let you know when the registration page is up. Our next intake is scheduled for February 2025.
Feedback from a previous iteration can be found here.
Key summary
- The new course is open and will run for 7 weeks. This course is online, but at your own pace, meaning that you need to dedicate enough time (5-8h per week) to fully benefit from the program.
- There are 4 weeks of asynchronous learning (you work at our own pace to cover the material each week).
- There are 4 live webinars organised on the first 4 Thursdays at 5pm AEST (convert your time here) to summarise some key concepts and ask your questions (the webinars will be recorded, as there are daylight savings during this period).
- You will have the opportunity to chat on Slack and ask your questions during the whole course.
- You can analyse your own data for the assessment (due in week 6) or use the data provided. You will reinforce your learning by marking the assignments of 2-3 other learners.
- Teaching Period Dates, asynchronised:
- Teaching commences: Monday, 21 Oct 2024, 9:00 am AEST
- Teaching concludes: Sunday, 17 Oct 2024, 11:59 pm AEST (after 4 weeks)
- (non marked) Assessment due: Friday 29 Nov 2024 (2 weeks prep)
- Peer-review of assessment due: Friday 6 Dec 2024 (1 week prep)
- Fees vary for
- Research Higher Degree students enrolled at a University: $495 AUD (incl. GST) [discount code: MIXO_RHD]
- Staff and members from Universities & Not-for-profit organisations: $825 (incl. GST) [discount code: MIXO_NFP_STAFF]
- Other industries: $1320 AUD (incl. GST)
- discounts of 5% for a group of 3-9 learners and 10% for 10+ learners, however, this will require a single invoice per group.
(these funds go towards the support of a software developer to maintain the package)
Information about the course and registration: https://study.unimelb.edu.au/find/short-courses/mixomics-r-essentials-for-biological-data-integration/
The number of places is limited, so first come first serve (this course runs once or twice a year)
What if I need an invoice? Contact Student Support at continuing-education[at]unimelb.edu.au
Prerequisites. A good working knowledge in R programming (e.g. handling data frame, perform simple calculations and display simple graphical outputs) is essential to fully benefit from the course*. The course is divided into theory (50%) and hands-on practice, with the opportunity to analyse your own data. The exercises and assignments are in R. Participants are encouraged to use RStudio and Rmarkdown (template and R code provided).
- For those looking for an R refresher well ahead of the course:
- https://monashdatafluency.github.io/r-intro-2/index.html
- the R cheatsheets for reference: https://iqss.github.io/dss-workshops/R/Rintro/base-r-cheat-sheet.pdf
*Learners who are not proficient in R do not get the full benenefit of the course (based on their own, honest, feedback!)
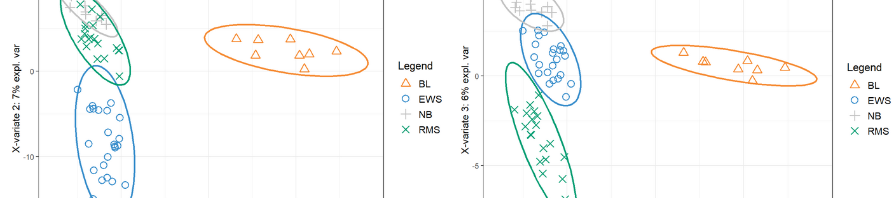
Webinar: Microbial network inference for longitudinal microbiome studies with LUPINE
Our latest method based on PLS to infer microbial networks across time is now in preprint!
- LUPINE is a PLS-based method that combines dimension reduction, and partial correlations to infer associations between taxa.
- LUPINE takes into account information across time points
- LUPINE has been designed for relatively small sample sizes and small number of time points
Microbial network inference for longitudinal microbiome studies with LUPINE. Saritha Kodikara, Kim-Anh Lê Cao. bioRxiv 2024.05.08.593086; accepted in Microbiome.
View this 50min video of Dr Saritha Kodikara presenting her method LUPINE:
We also have a second video presented by Prof Kim-Anh Lê Cao who sets LUPINE in the context of microbiome longitudinal data analysis, elaborating more on the types of analytical objects covered in Kodikara et al. (2022) Statistical challenges in longitudinal microbiome data analysis, Briefings in Bioinformatics.
Below you will also find the most common questions related to LUPINE.
FAQ:
Q: Do you build up the network from the covariance matrix or from the inverse covariance matrix? And what are you doing linear regression on?
A: The network is built on the partial correlation so it would be similar to the inverse covariance matrix. But instead of estimating the inverse covariance matrix, we calculate partial correlations through linear regression. To estimate the partial correlation between taxa a and taxa b, we regress their counts on the low dimensional representation of other taxa (excluding taxa a and b). This is then repeated for all pairs (we have an efficient way to do this computationally).
Q: You reduce the dimension of the data into one dimension. How much variance can be explained by the 1st component in your computation?
A: It depends on the data, but in the data we analysed, and if consider the single time point scheme only with PCA, the first component explained about 25% of the total variance. We could add more components into the regression but that may overfit the regression model. This is why we only select the first component, which explains much of the variance (for PCA, single time point) or covariance (for PLS, multiple time points).
Q: Do you think that this approach would work on single cell data trying to look at gene co expression in sort of longitudinal data in across time points?
A: It will not work with the present single cell technologies, because in LUPINE we need the same individuals/samples/cells across time to infer the association.
Q: When you do the linear regression, do you regress directly on the counts with all the zeros and the sparsity that you mentioned?
A: Yes, the method was originally developed for count data. We regress on the count data, but we also include library size as an offset to account for different library sizes. The method also works with center log ratio values, which I used to analyse the third case study.
Q: Do you apply your method for the two groups combined or separately?
A: I model each group separately as we assume that each group has a unique network.
Q: You’re building the networks building based on the partial correlations. What about the actual network for representation, do you actually binarize it?
A: Yes, I binarize the network based on a correlation test.
The LUPINE package
LUPINE is currently not directly available from the mixOmics package, instead it is a separate R package that can be installed from Github.
[closed] Self-paced online course Feb 5 – March 22 2024
This workshop is now closed. Fill in this short survey to register your interest. A new iteration of the course might be run between Sept – Nov if there is sufficient interest!
Key summary
- The new course is open and will run for 7 weeks. This course is online, but at your own pace, meaning that you need to dedicate enough time (5-8h per week) to fully benefit from the program.
- There are 4 weeks of asynchronous learning (you work at our own pace to cover the material).
- There are 4 live webinars organised on the first 4 Thursdays at 5pm AEST (convert your time here) to summarise some key concepts and ask your questions (the webinars will be recorded).
- You will have the opportunity to chat on Slack and ask your questions during the whole course.
- You can analyse your own data for the assessment (due in week 6) or use the data provided. You will reinforce your learning by marking the assignments of 2-3 other learners.
Feedback from the 2022 iteration can be found here.
- Teaching Period Dates, asynchronised:
- Teaching commences: Monday, 5 Feb 2024, 9:00 am AEST
- Teaching concludes: Sunday, 29 Feb 2024, 11:59 pm AEST (4 weeks)
- (non marked) Assessment due: Friday 15 March 2024 (2 weeks)
- Peer-review of assessment due: Friday 22 March 2024 (1 week)
- Fees vary for
- Research Higher Degree students enrolled at a University: $495 AUD (incl. GST) [discount code: MIXO_RHD]
- Staff and members from Universities & Not-for-profit organisations: $825 (incl. GST) [discount code: MIXO_NFP_STAFF]
- Other industries: $1320 AUD (incl. GST)
- discounts of 5% for a group of 3-9 learners and 10% for 10+ learners, however, this will require a single invoice per group.
(these funds go towards the support of a software developer to maintain the package)
- Information about the course and registration:
- Information about the course: https://study.unimelb.edu.au/find/short-courses/mixomics-r-essentials-for-biological-data-integration/
- Registration: https://purchase.study.unimelb.edu.au/mixomics-r-essentials-for-biological-data-integration-sc-b2c-open-online-feb24-1-prog-mixomics-r-essentials-for-b-sc-b2c-open-online-feb24-1-0001
The number of places is limited, so first come first serve (this course runs once or twice a year)
What if I need an invoice? Contact Student Support at continuing-education[at]unimelb.edu.au
Prerequisites. A good working knowledge in R programming (e.g. handling data frame, perform simple calculations and display simple graphical outputs) is essential to fully benefit from the course*. The course is divided into theory (50%) and hands-on practice, with the opportunity to analyse your own data. The exercises and assignments are in R. Participants are encouraged to use RStudio and Rmarkdown (template and R code provided).
- For those looking for an R refresher well ahead of the course:
- https://monashdatafluency.github.io/r-intro-2/index.html
- the R cheatsheets for reference: https://iqss.github.io/dss-workshops/R/Rintro/base-r-cheat-sheet.pdf
*Learners who are not proficient in R do not get the full benefit of the course (based on their own, honest, feedback!)
[closed] Self-paced online course May 22 – July 7 2023
If you’ve missed out, our next iteration will run from 19th Feb – 5th April 2024. You can fill up this short survey to be notified when we open our next course.
Summary
- The new course is open and will run for 7 weeks. This course is online, but at your own pace, meaning that you need to dedicate enough time (5-8h per week) to fully benefit from the program.
- There are 4 weeks of asynchronous learning (you work at our own pace to cover the material).
- There are 4 live webinars organised on Thursdays at 5pm AEST (convert your time here) in the first 4 weeks to summarise some key concepts and ask your questions (the webinars will be recorded).
- You will have the opportunity to chat on Slack and ask your questions during the whole course.
- You can analyse your own data for the assessment (due in week 6) or use the data provided. You will reinforce your learning by marking the assignments of 2-3 other learners.
Feedback from the 2022 iteration can be found here.
- Teaching Period Dates, asynchronised:
- Learning Start: Monday, 22 May 2023, 9:00 am AEST
- Learning Ends: Sunday, 18 June 2023, 11:59 pm AEST (4 weeks)
- (non marked) Assessment due: Friday 30th June 2023 (2 weeks)
- Peer-review of assessment due: Friday 7th July 2023 (1 week)
- Fees vary for
- Research Higher Degree students enrolled at a University: $495 AUD (incl. GST)
- Staff and members from Universities & Not-for-profit organisations: $825 (incl. GST)
- Other industries: $1320 AUD (incl. GST)
- discounts of 5% for a group of 3-9 learners and 10% for 10+ learners, however, this will require a single invoice per group.
(these funds go towards the support of a software developer to maintain the package)
- Information about the course and registration:
- Information about the course: https://study.unimelb.edu.au/find/short-courses/mixomics-r-essentials-for-biological-data-integration/
- Registration: https://your.unimelb.edu.au/s/event/a2a2e0000008buM/mixomics-r-essentials-biological-may-23-b2c
The number of places is limited, so first come first serve (we aim to run this course twice a year).
What if I need an invoice? Contact Student Support at continuing-education[at]unimelb.edu.au
Prerequisites. A good working knowledge in R programming (e.g. handling data frame, perform simple calculations and display simple graphical outputs) is essential to fully benefit from the course*. The course is divided into theory (50%) and hands-on practice, with the opportunity to analyse your own data. The exercises and assignments are in R. Participants are encouraged to use RStudio and Rmarkdown (template and R code provided).
- For those looking for an R refresher well ahead of the course:
- https://monashdatafluency.github.io/r-intro-2/index.html
- the R cheatsheets for reference: https://iqss.github.io/dss-workshops/R/Rintro/base-r-cheat-sheet.pdf
*Learners who are not proficient in R do not get the full benefit of the course (based on their own, honest, feedback!)