- Welcome
- Workshops
- [open] Self-paced online course Feb 24 – April 11, 2025
- [closed] Self-paced online course Oct 21 – Dec 6 2024
- Self-paced online course Feb 19 – April 5 2024
- [open] Self-paced online course May 22 – July 7 2023
- [closed] 13-14 March 2023, Brisbane, Aus
- Self-paced online course Oct 31st – Nov 27 2022
- [Closed] Self paced online course Oct 11 – Nov 7 2021
- [closed] Online workshop (on-demand)
- [cancelled] 26-29 Oct 2021, Palmerston North, NZ (beginner)
- Workshops for 2019 – 2020
- 12-13 March 2020, Advanced workshop, L’Oréal headquarters
- Feb 3-5 2020, Perth, AUS (beginner, omics and microbiome)
- Software requirements for 2020 mixOmics workshops
- June 4-6 2019, Toulouse, FR (beginner, 3 days)
- April 15 – 17 2019, Melbourne AUS (beginner, microbiome)
- Nov 6 2018, Vancouver (microbiome)
- Beginner workshop 23-25 July 2018, Melbourne
- July 8 2018, Barcelona, SP (Introductory)
- June 7-8 June 2018, Saclay, FR (advanced)
- April 12-13 2018, Sydney, AUS
- Software requirements for mixOmics workshops
- Nov 22-24 2017, Toulouse, FR
- Nov 9-10 2017, Toulouse, FR
- Oct 23-24 2017, Toulouse, FR, Advanced Workshop
- Nov 4 2016, Brisbane, AUS
- Sept 12-14 Sept 2016, Toulouse, FR
- 22 July 2016, Saclay, FR Summer School Saclay Plant Sciences
- July 11-13 2016, Clermont-Ferrand, FR
- March 24-25 2016, Nantes, FR
- Sept 24-25 2015, Jouy-en-Josas, FR
- Sept 14-15 2015, Toulouse, FR
- Sept 10-11 2015, Montpellier, FR
- Aug 13-14 2015, Brisbane, AUS
- April 9-10 2015, Auckland, NZ
- October 6-7 2014, Toulouse, FR
- Sept 7, 2014, Strasbourg, FR
- Book
- Webinars
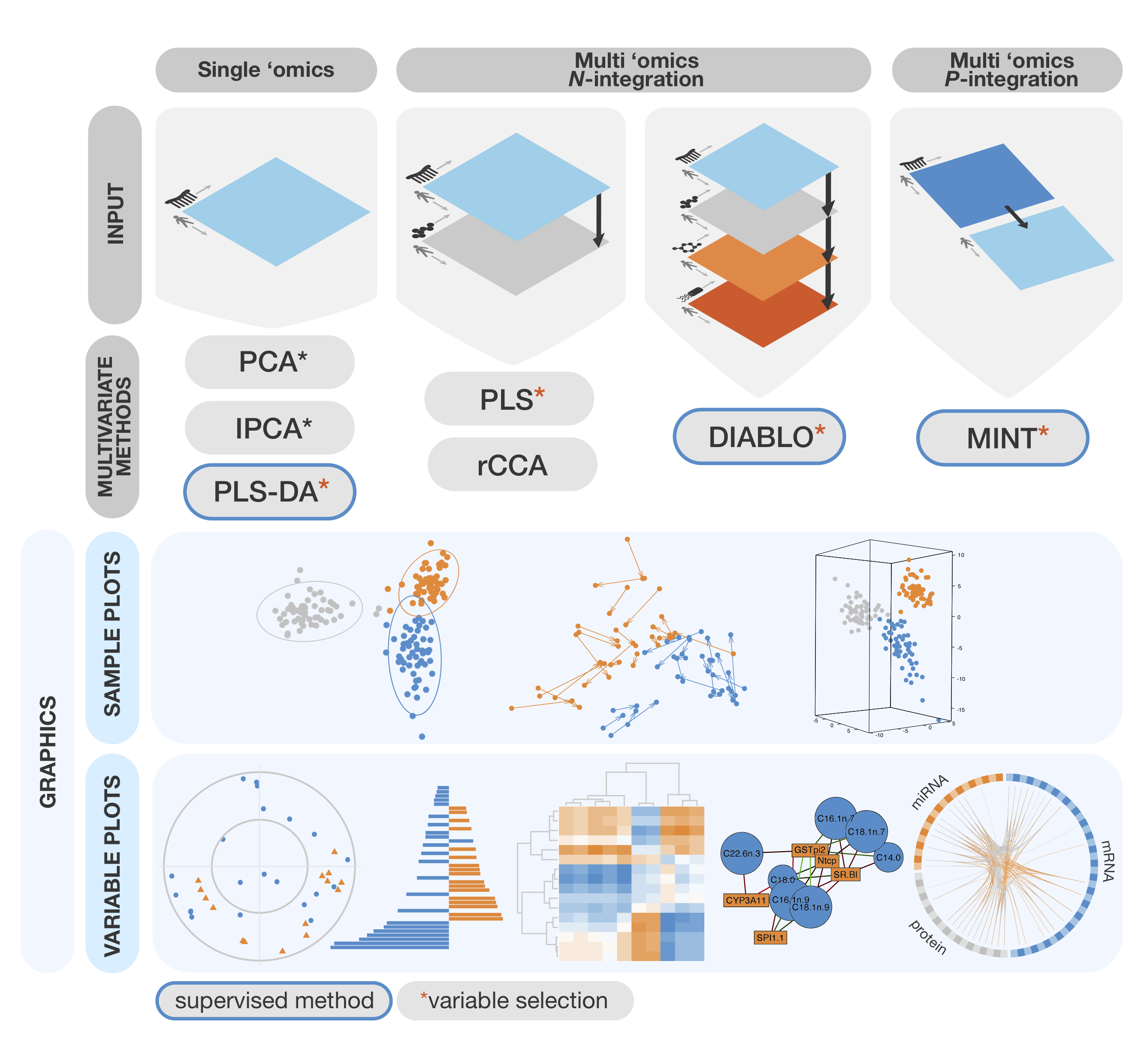
- Methods
- Graphics
- Case Studies
- FAQ
- About