R CRAN update
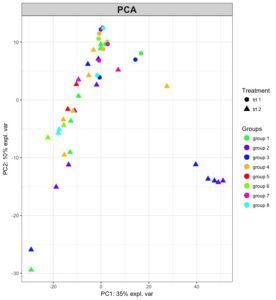
The new patch version of mixOmics is on CRAN. It includes a few bug fixes raised by our users (thank you!) and a few improvements. Florian Rohart has been fiddling really hard with ggplot2 to make a new plotIndiv version that can beautifully handle two legends!
# indicate the group, treatment and pch for each sample
my.group
[1] "group 1" "group 1" "group 2" "group 2" "group 3" "group 3" "group 4" "group 4" "group 1" "group 1" "group 2" "group 2" "group 3" "group 3" "group 4" "group 4"
[17] "group 1" "group 1" "group 2" "group 3" "group 3" "group 4" ....
my.treatment
[1] "trt 2" "trt 2" "trt 2" "trt 2" "trt 2" "trt 2" "trt 2" "trt 2" "trt 2" "trt 2" "trt 2" "trt 2" "trt 2" ....
my.pch.trt
[1] 17 17 17 17 17 17 17 17 17 17 17 17 17 17 17 17 16 16 ....
plotIndiv(pca.res, ind.names = F, title = 'PCA', legend = TRUE,
# legend 1 colors setting:
group = my.group, col.per.group = color.per.group, legend.title = 'Groups',
# pch setting:
legend.title.pch = 'Treatment', pch = my.pch.trt, pch.levels = my.treatment)
Here is a list of the major bug fixes and improvements for 6.1.2:
New features:
————-
1 – tune.splsda now returns a ‘choice.ncomp’ which indicates the number of components to choose (only if nrepeat > 2, criterion based on t-tests)
2 – plotIndiv now enables two legends based on color, as well as pch, when pch is a factor different from what is indicated in group (use arguments pch and pch.levels, see ?plotIndiv)
Enhancements:
————-
1 – argument ‘cutoff’ now replaces ‘threshold’ in network for consistency with plotVar and circosPlot
2 – new argument ‘sd’ in plot.perf for block.splsda method
3 – new arguments “color.Y” and “color.blocks” in cimDiablo
4 – new argument ‘xlim’ in plotLoadings
Bug fixes:
———-
– directionality is now enforced in AUROC (results lower than 0.5 can be obtained, which would indicate a very poor model performance)
Manuscripts:
The MINT paper is out:
-
Rohart F., Matigian N., Eslami A., Bougeard S and Lê Cao, K. A.MINT: A multivariate integrative method to identify reproducible molecular signatures across independent experiments and platforms. Now available on bioRxiv! in press in BMC Bioinformatics 18:128.
The mixOmics manuscript (first draft) is on bioRxiv, with sweave codes:
- Rohart F., Gautier, B, Singh, A and Lê Cao, K. A. mixOmics: an R package for ‘omics feature selection and multiple data integration. On bioRxiv. Sweave and R scripts available here.